Two constraint-based algorithms are available for structure learning: The PC algorithm and the NPC algorithm. In this tutorial we shall assume that you already known how these algorithms work.
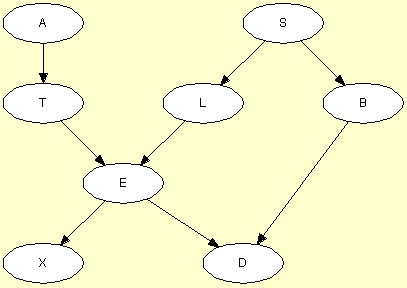
Consider the data file asia.dat sampled from the Chest Clinic network shown in Figure 1.

Figure 2: The structure from which the data were sampled.
Figure 2 shows the first few lines of the data file, which consists of 10000 cases altogether.
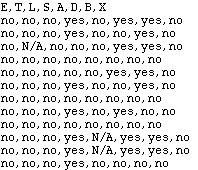
The structure learning functionality is available under the "File" menu and through the structure learning button of the tool bar of the Main Window. The structure learning button is shown in Figure 3.
When the button is pressed, the structure learning dialog appears (see Figure 4). Note the field "Level of Significance" which specifies the level of significance for the statistical independence tests performed during structure learning. Press the "Select File" button and choose a file from which the structure is to be estimated. When a file has been selected the "OK" button gets enabled.
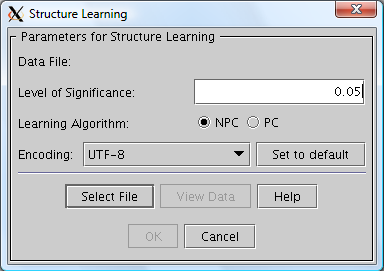
Now, before activating the OK button, you should select which learning algorithm you wish to use: The PC algorithm or the NPC algorithm. The descriptions of these algorithms are also available through the Help button of the structure learning dialog (see Figure 4).
Pressing the "OK" button starts the selected structure learning algorithm.
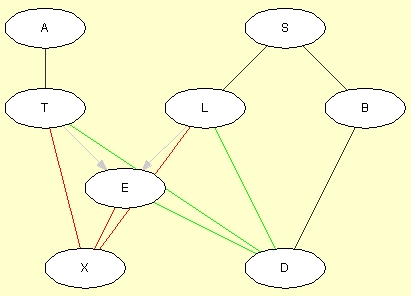
If the PC algorithm was selected, the result should appear as in Figure 5. Obviously, the structure of the original network has not been recalled perfectly. The problem is that variable E depends deterministically on variables T and L through a logical OR (E stands for Tuberculosis or Lung cancer). This means that (i) T and X are conditionally independent given E, (ii) L and X are conditionally independent given E, and (iii) E and X are conditionally independent given L and T. Thus, the PC algorithm concludes that there should be no links between T and X, between L and X, and between E and X. Obviously, this is wrong. The same reasoning leads the PC algorithm to leave D unconnected to T, L, and E. Also, as the PC algorithm directs links more or less randomly (respecting, of course, the conditional independence and dependence statements derived from data), the directions of some of the links are wrong.
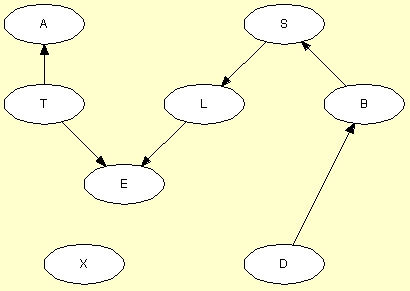
Figure 5: The structure learned by the PC algorithm.
If the NPC algorithm was selected and there are uncertain links or links for which the directionality could not be determined for sure, the user will be presented with an intuitive graphical interface for resolving these structural uncertainties (see the description of the NPC algorithm for details). Figure 6 shows the (intermediate) result of the NPC structure learning algorithm applied to the same data.
Depending on the choices made by the user, many different final structures (including the original structure shown in Figure 1) can be generated from this intermediate structure.
The Hugin Graphical User Interface implements a number of different algorithms for learning the structure of a Bayesian network from data, see Structure Learning for more information.
Once the structure of the Bayesian network has been generated, the conditional probability distributions of the network can be estimated from the data using the EM-learning algorithm, see the EM tutorial.